Research Groups
Research of our groups is centerd on the integrative structural analysis of macromolecular complexes in genome biology.
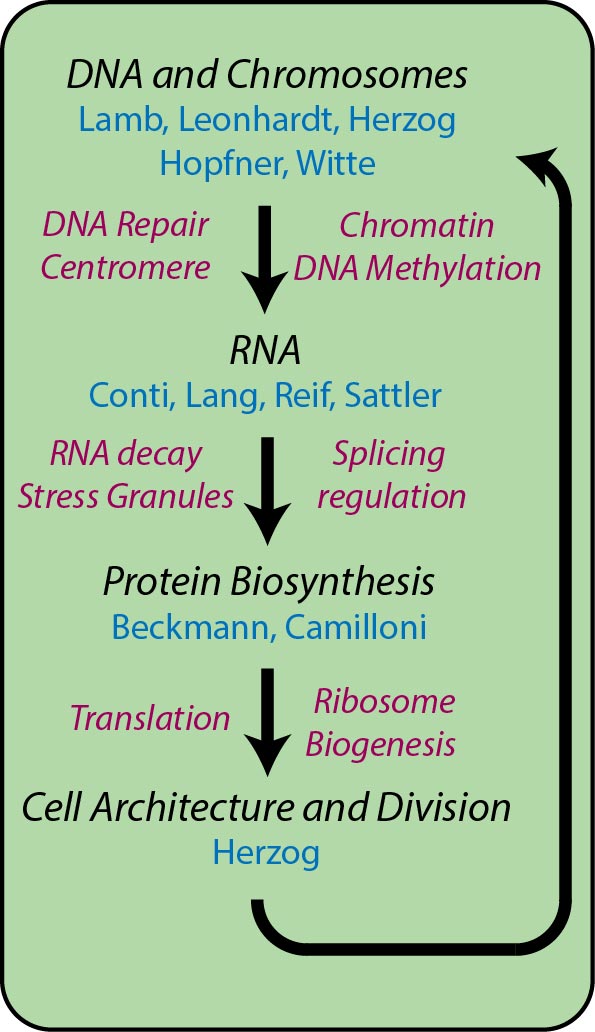
They include research topics like:
- DNA- and chromosome-associated processes (DNA repair, centrosome organization and DNA modification: A4 Hopfner, A10 Witte, C1 Leonhardt, C3 Lamb, C5 Herzog)
- RNA-associated processes (RNA decay, stress granules, splicing: A2 Conti, A5 Sattler, A11 Reif, C4 Lang)
- processes involved in protein biosynthesis (translation, ribosomes, protein folding: A1 Beckmann)
- processes controlling cell division (C5 Herzog)
Research Area A (Structural Protein Science and Hybrid Methods)
- A1 Roland Beckmann: Molecular architecture of a pre-60S ribosomal particle from S. cerevisiae
- A2 Elena Conti: Structure and function of deadenylation complexes in eukaryotic RNA decay
- A4 Karl-Peter Hopfner: Dynamic architecture of the DNA double-strand break repair and resection machinery
- A5 Michael Sattler: Dynamics and RNA recognition by multi-domain RNA-binding proteins in splicing regulation
- A10 Gregor Witte: Regulation of the DNA-damage checkpoint protein DisA bound to Holliday junctions by the AAA+-protein RadA
- A11 Bernd Reif: Structural analysis of TIA-1 induced Stress Granules
Research Area C (Advanced Imaging and Crosslinking Methods)
- C1 Heinrich Leonhardt: Dynamics of protein complexes involved in DNA demethylation by base excision repair
- C3 Don Lamb: Investigating DNA double-strand break repair using single-molecule fluorescence microscopy and DNA Origami
- C4 Kathrin Lang: New chemical tools for structural biology
- C5 Franz Herzog: Identifying the MAD1 Receptor at the Kinetochore and its Role in Checkpoint Signaling using a Hybrid Approach

